
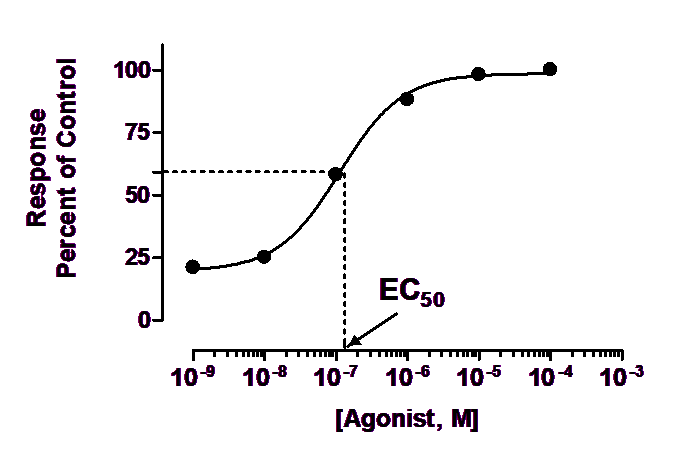
Drug cytotoxicity and Bliss Synergy were analyzed using GraphPad Prism and Combenefit. Patient-derived tumor organoids were tested for sensitivity to drugs matched to the predicted targets as single agent or in combination, using a 4-fold dilution followed by 72h viability assay. Methods: Following PATRIOT analysis, patients with predicted CDKN2A downregulation or MET overexpression consisting primarily of colon and pancreatic cancer were grouped into two cohorts. To further validate this platform, we interrogated the predictive efficacy of PATRIOT in cohorts of patients characterized by CDKN2A downregulation or MET overexpression. Patient-derived tumor organoids showed a significant response to the suggested treatment, Olaparib, supporting the reliability of PATRIOT prediction. In our previous study, we tested the PATRIOT platform on a patient with predicted KRAS and IDHI inactivation resulting in inhibition of DNA damage repair pathway.
We have previously described our PATRIOT platform, which integrates RNA DE analysis, NGS, and high throughput patient-derived organoid models to identify new therapeutic targets. With the development of next-generation sequencing, the field of precision oncology has been expanded to tailor treatments to a patient's mutational profile, however, there is a need for additional information such as RNA Differential Expression (DE) to match more patients to potentially beneficial drugs. ) + theme ( = element_blank ( ), legend.title = element_blank ( ), legend.position = c ( 0.05, 0.95 ), legend.justification = c ( 0.05, 0.Background: Cancer therapy has witnessed great progress over the last 50 years with advancements in diagnostic tools to enable more precise treatment. # construct the ame, log10 transform the agonist concentration # convert the ame to long format, then remove any rows with NA df % mutate (log.agonist = log10 ( agonist ) ) %>% pivot_longer ( c ( - agonist, - log.agonist ), names_pattern = "(.


 0 kommentar(er)
0 kommentar(er)
